OnePot Pro DNA Library Prep Kit for Illumina 24T
Description
OnePot Pro DNA Library Prep Kit for Illumina is a new generation enzymatic fragmentation-based library prep kit professionally developed for Illumina high-throughput sequencing platform. This set simplifies the operation process compared with the traditional library construction method and also greatly reduces the time and cost by performing fragmentation, end repair and dA-tailing of dsDNA in one reaction. This library prep kit has an excellent library conversion rate and is applicable for samples from all common animals, plants, microorganisms, etc., and also the FFPE samples. This upgraded kit using the latest optimized ligase greatly decreases the self-ligation rate during adapter ligation. Moreover, the introduction of a new high-fidelity polymerase further improves the homogeneity and fidelity of amplification.
Features
- Applicable to 500 pg - 1 μg genomic DNA, full-length cDNA, FFPE DNA samples and other samples
- High-quality fragment enzyme, which can randomly cut double-stranded DNA, and has no preference for cutting fragments
- Fragmentation, end repair/adding A in one step
- High-fidelity enzyme with strong amplification efficiency, significantly improving library quality and yield
- Strict batch performance and stability quality control
Applications
- Construction of DNA/cDNA library for illumina platform
- Suitable for sequencing in tumor, reproductive genetics, genetic disease detection and other fields
- PCR-free library construction
Work flow

Specifications
| Product Type | Library Preparation Kit |
| Libraries | Fragment Library |
| Fragmentation method | Enzyme |
| Input amount | 100 pg - 1000 ng |
| For Use With (Equipment) | Illumina Platforms |
| Sample Type | gDNA、DNA |
| Sequencing Type | Genome & DNA Sequencing |
| Product Line | DNA library construction |
| For Use With (Application) | NGS library preparation |
| Quantity | 24/96 Reaction |
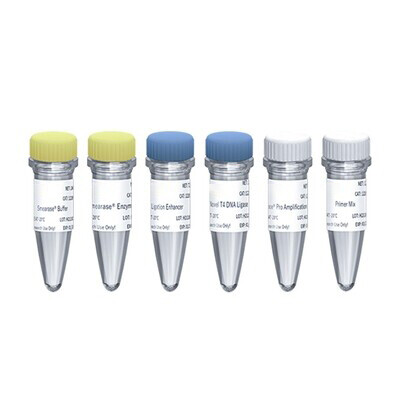
Components
| Components No. | Components | 12205ES24 |
| 12205-A | SmearaseTM Buffer | 240 μL |
| 12205-B | SmearaseTM Enzyme | 120 μL |
| 12205-C | Ligation Enhancer | 720 μL |
| 12205-D | Novel T4 DNA Ligase | 120 μL |
| 12205-E | CanaceTM Pro Amplification Mix | 600 μL |
| 12205-F | Primer Mix | 120 μL |
Shipping and Storage
Dry ice shipping. -15℃ ~ -25℃ storage, valid for one year.
Figures
- Enzyme fragmentation performance

Figure 1. Library construction of different sample types with Cat#12205
Sample: Human Blood gDNA, human saliva gDNA and Bacillus cereus
Method: Enzyme digestion time:12 minute, Aligent 2100 detect the range of enzyme digestion fragments
Results: The distribution of the library was basically consistent with different samples and different amounts of enzyme digestion for 12 minutes.

Figure 2. Different template input compatibility test with Cat#12205
Sample: Human Blood gDNA 20ng, 50ng, 100ng, 500ng, 1ug;
Method: Enzyme digestion time:12 minute, Aligent 2100 detect the range of enzyme digestion fragments
Input amount of different formworks (20ng~1μg) The Results: the size of the digested products are consistent, indicating that the amount of template input will not affect the size of the digested fragments
- GC Bias test performance

Figure 3. Sequencing data analysis GC Bias results
Three bacterial strains with different GC contents of ZYMO standard D6306 were tested with Cat#12205 kit.
The sequencing results showed that the kit had no bias for strains with different GC contents.